Background: Achieving undetectable minimal residual disease (MRD) has been considered a functional cure in multiple myeloma (MM), emphasizing the clinical importance of depth and duration of remission. Interestingly, a subset of myeloma patients presents with an MGUS-like status, characterized by persistent M-component or detectable MRD despite systematic treatment. Surprisingly, these patients exhibit an indolent disease course and experience unexpectedly long survival (PFS ≥ 5 years). Nevertheless, the specific genomic characteristics of their tumor cells, immune microenvironment, and clonal evolution patterns are still inadequately explored.
Methods: This study utilizes data from the National Longitudinal Cohort of Hematological Diseases in China (NCT04645199) to investigate the unique MGUS-like cohort. This cohort comprises patients who achieved functional cure (PFS ≥ 5 years) but retained MGUS-like features. We conducted DNA and RNA sequencing of sorted plasma cells to elucidate the specific transcriptional and genomic profiles and clonal evolution patterns, aiming to shed light on the underlying mechanisms and enable individualized treatment approaches.
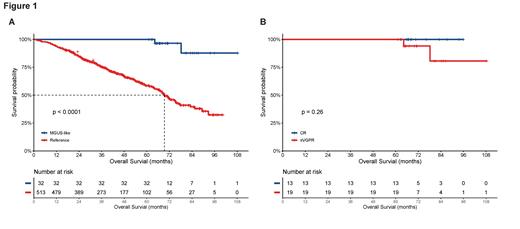
Results: In the study cohort, only 6.4% (35/548) exhibited the MGUS-like profile, characterized by younger age, low tumor burden, infrequent anemia and bone lesions, and stable cytogenetics. Intriguingly, the MGUS-like cohort demonstrated consistently favorable prognoses regardless of response depth, unlike classical MM patients. Analysis of the intrinsic features of tumor cells revealed a distinct transcriptome for the MGUS-like group compared to classical MM. Subsequently, we identified a MGUS-like transcriptional gene signature that effectively discriminated a rare cohort with excellent survival, irrespective of remission depth, in the MMRF database. Notably, gene set enrichment analysis (GSEA) analysis revealed the upregulation of genes involved in the adaptive immune response, T cell activation, natural killer cell-mediated immunity, and erythrocyte differentiation, along with the downregulation of genes involved in protein folding. These biological differences potentially account for the unprecedented favorable prognosis due to an activated immune microenvironment and distinct clinical features such as reduced anemia and low tumor burden. Furthermore, genomic analysis of sequential samples from MGUS-like patients demonstrated that most mutations and copy number alterations became undetectable in residual tumor cells or during relapse, suggesting clonal selection of indolent tumor clones in MGUS-like patients.
Conclusions: The rare MGUS-like population exhibits unique clinical features and an exceptionally favorable prognosis with limited treatment. Utilizing the distinct tumor transcriptome and specific clonal selection pattern, early identification of MGUS-like patients could facilitate precise individualized treatment strategies.
Disclosures
No relevant conflicts of interest to declare.